Research
Equivalence between Interpretable Neural Network and Kernel Logistic Regression
The work of my thesis aimed to develop an explainable supervised classification method, thereby meeting the requirements of precision medicine. This research falls within the fields of Explainable Artificial Intelligence (XAI) and neural network learning.
I developed an interpretable neural network called SATURNN (Splines Approximation Through Understandable ReLU Neural Network), whose scoring function is modeled as an additive sum of univariate spline functions [Article].
I developed an interpretable neural network called SATURNN (Splines Approximation Through Understandable ReLU Neural Network), whose scoring function is modeled as an additive sum of univariate spline functions [Article].
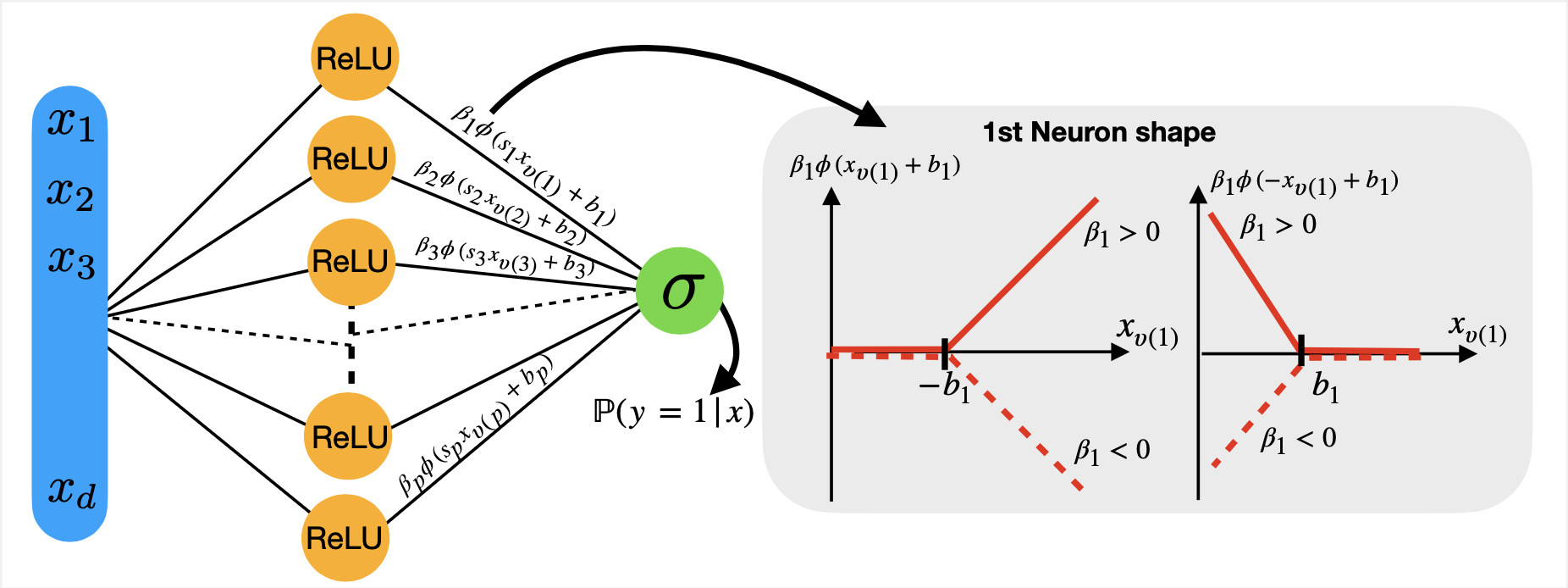
This model extends interpretable paradigms such as Generalized Additive Models (GAMs) and Neural Additive Models (NAMs). However, although interpretable, SATURNN, like any neural network, does not guarantee convergence to a unique solution.
To address this limitation, we linearized the SATURNN scoring function in infinite dimension (comprising a large number of neurons) in the neighborhood of its initializations, thus showing that this network can be approximated by logistic regression applied to data transformed by this linearized function. However, the performance of this approach is influenced by the randomness of SATURNN’s initializations.
Finally, we demonstrated that this linearized scoring function can be reformulated as a kernel, asymptotically converging to a finite limit. This led to the creation of the EKLR (Expected Kernel Logistic Regression) method, in which the variable segmentation by the kernel is deterministic and does not require any additional learning parameters. The resulting decision rule is an additive sum of univariate splines, interpretable and accompanied by convergence guarantees [Article].
Keywords: Neural Networks, Logistic Kernel Regression, Tangent Neural Kernel, Generalized Additive Models, Univariate Splines, Interpretable Machine Learning, Explicable Artificial Intelligence, Precision Medicine.
Representation Learning on SNDS reimbursement data
As part of my postdoctoral work, my research aims to develop a decision-support algorithm for oncologists based on the analysis of data from health insurance centralized in the Système National des Données de Santé (SNDS). This vast repository of Electronic Health Records (EHRs) allows the tracking of patients’ therapeutic pathways, represented as temporal sequences of visits. These unstructured, high-dimensional data present major methodological challenges. I first focused on evaluating Representation Learning methods for patients, which consist of projecting EHR information into a reduced-dimensional latent space. Our work revealed that the most effective unsupervised deep Representation Learning approaches in terms of common empirical metrics fail to accurately represent the clinical reality [Article].
Furthermore, given the access constraints to SNDS data (special training and authorizations), I collaborated with Thomas Guyet (INRIA Lyon) to simulate a realistic breast cancer database . This methodological work, based on carefully calibrated probabilistic laws, faithfully reproduces patients’ therapeutic pathways while respecting confidentiality requirements. This work (in submission) makes the simulated database freely available, reflecting the architecture and specificities of SNDS data and thus enabling practice on this data as well as algorithm development and evaluation.
Finally, due to its highly complex architecture, high dimensionality, and the abstract, very specific coding of medical events, the SNDS is difficult to exploit. To address this challenge, we developed a Python package , pySNDS, which simplifies the identification and interpretation of relevant information in this complex database, thus facilitating its use for research purposes. This package is currently under submission.
Furthermore, given the access constraints to SNDS data (special training and authorizations), I collaborated with Thomas Guyet (INRIA Lyon) to simulate a realistic breast cancer database . This methodological work, based on carefully calibrated probabilistic laws, faithfully reproduces patients’ therapeutic pathways while respecting confidentiality requirements. This work (in submission) makes the simulated database freely available, reflecting the architecture and specificities of SNDS data and thus enabling practice on this data as well as algorithm development and evaluation.
Finally, due to its highly complex architecture, high dimensionality, and the abstract, very specific coding of medical events, the SNDS is difficult to exploit. To address this challenge, we developed a Python package , pySNDS, which simplifies the identification and interpretation of relevant information in this complex database, thus facilitating its use for research purposes. This package is currently under submission.
Keywords : Electronic Health Records, Deep Representation Learning, Système National des Données de Santé.